Notes to self. Often, discussions related to maximum likelihood refer to (A) estimating parameters likely to have come from some (B) probability distribution (PD) by (C) maximizing a likelihood function (from Wikipedia).
In many cases, (A) is the unknown which we want to figure out (e.g., means, variances, regression coefficients). We make an assumption about the PD where the data come from and determine the likelihood or the log likelihood of observing the datum. Get the product of all the likelihoods (multiplicative) or the sum of the log likelihoods (additive) and you will get the total likelihood.
An example will help:
Simulate some data
set.seed(123)
x <- rnorm(10, mean = 110, sd = 3) #make up some data
x## [1] 108.3186 109.3095 114.6761 110.2115 110.3879 115.1452 111.3827 106.2048
## [9] 107.9394 108.6630mean(x)## [1] 110.2239sd(x)## [1] 2.861352We have a vector with 10 elements. We can easily get the mean of this by just typing mean(x) but this is for illustrative purposes.
We create a likelihood function llike based on a normal distribution using:
\(f(x; \mu, \sigma^2) = \frac{1}{\sigma \sqrt{2\pi}}e^{-\frac{1}{2}(\frac{x - \mu}{\sigma})^2}\)
This take the sums of the log likelihood of whatever data is in x with a specified mean (mu) and standard deviation sigma– the later two we “don’t” know. There is also the built-in function of dnorm which we can use to check if we are getting the same results.
llike <- function(x, mu, sigma){
sum(log((1 / (sigma * sqrt(2 * pi))) * exp(-.5 * ((x - mu) / sigma)^2)))
}
llike(x, 100, 10) #testing function to see if this works## [1] -37.81005#just checking if we specify m = 100, sd = 10
sum(dnorm(x, mean = 100, sd = 10, log = T)) #using built-in function## [1] -37.81005We get the same result using a specified mean of 100 and an SD of 10.
We can take a series of means (mu below) and see where the sum of the ll is at the highest.
mu <- seq(100, 120, .1) #set of values to test
ll <- sapply(mu, function(z)
llike(x, z, 10) #find the log likelihood
)
plot(ll ~ mu) #plot 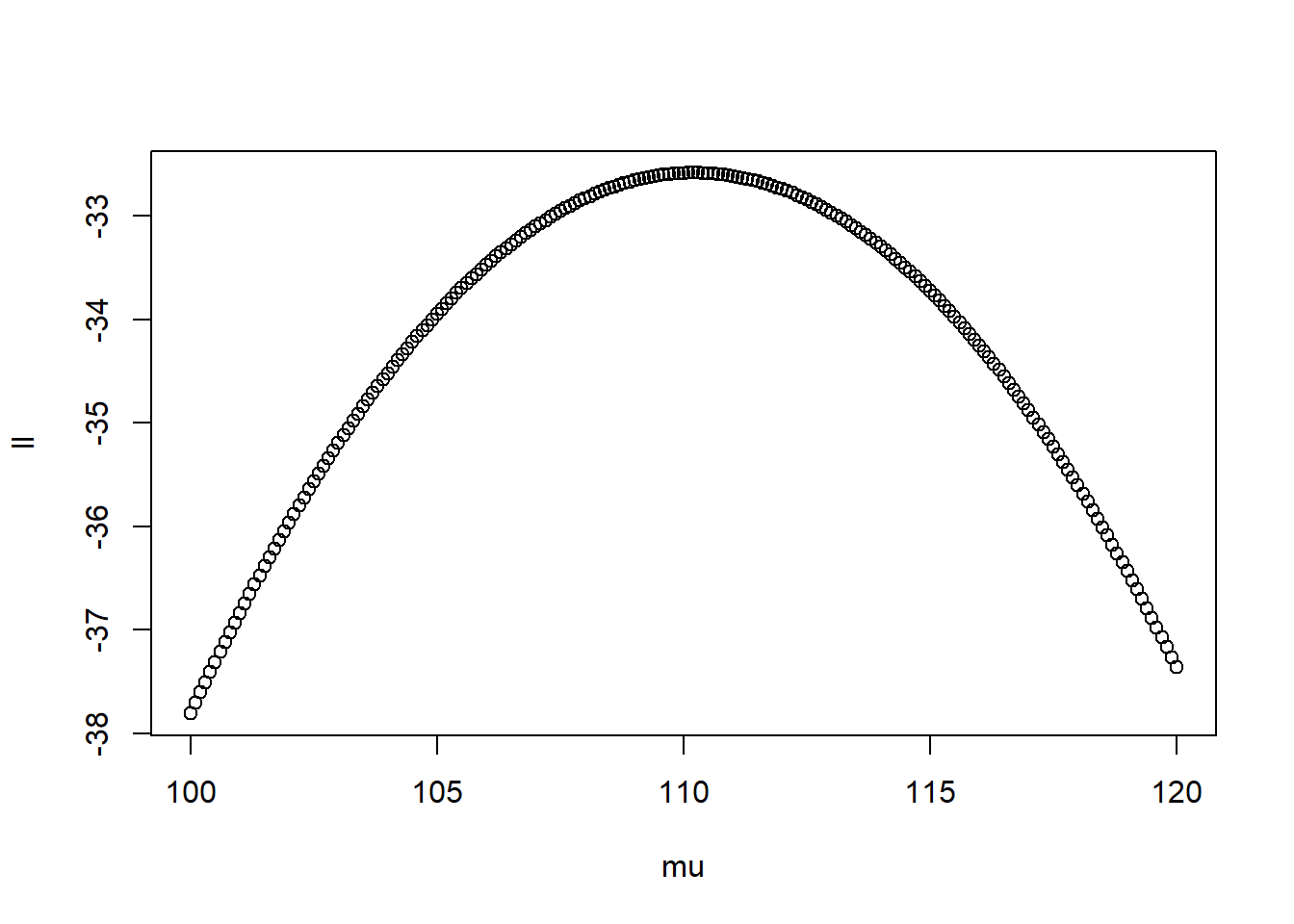
mu[which(ll == max(ll))] #which mu had the highest point## [1] 110.2Based on the results, it peaks around 110.
We can use another function to find the maximum point (optimizing involves either finding the maximum or minimum). A lot of these ideas come from the very useful and accessible book of Hilbe and Robinson (2013) Methods of Statistical Model Estimation.
## another function to optimize
bb <- function(p, x){
sum(dnorm(x, mean = p[1], sd = p[2], log = T))
}
## finding the maximum likelihood using 'optim'
fit <- optim(c(mean = 100, sd = 10), #starting values or guesses
fn = bb, x = x,
control = list(fnscale = -1)) #-1 to maximize
fit$par #contain the mean and the SD## mean sd
## 110.224295 2.715141mean(x)## [1] 110.2239sd(x) #this is the actual variance## [1] 2.861352sqrt(sum((x - mean(x))^2) / length(x)) #this is the ML variance## [1] 2.714517# (SS / n), not n - 1
# based on ML
# the variance is under estimatedWhat about using ML in a regression?
I have not seen examples showing this. I’ve only seen examples of the first kind shown (e.g., getting the mean).
First, create some data with two variables (x1 and x2):
## simulate some data
set.seed(2468)
ns <- 1000
x1 <- rnorm(ns)
x2 <- rnorm(ns)
e1 <- rnorm(ns)
y <- 10 + 1 * x1 + .5 * x2 + e1
dat <- data.frame(y = y, x1 = x1, x2 = x2)Prepare the formula and some matrices:
fml <- formula('y ~ x1 + x2')
df <- model.frame(fml, dat)
y <- model.response(df)
X <- model.matrix(fml, df)Create another function to optimize:
## log likelihood for continuous variables
ll.cont <- function(p, X, y){
len <- length(p)
betas <- p
mu <- X %*% betas
sum((y - mu)^2) #minimize this, sum of squared residuals
}In this case, we want to minimize the sums of squared residuals (error)– that’s what the best fitting line does.
start <- ncol(X) #how many start values
fit <- optim(p = rep(1, start), #repeat 1
fn = ll.cont,
X = X, y = y) #this minimizes the SSR
fit$par #compare with results below ## [1] 9.9952812 1.0151535 0.5098497## values are for the intercept / Bx1 / Bx2Compare to using our standard lm function:
t1 <- lm(y ~ x1 + x2, data = df)
summary(t1)$coef## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 9.9950936 0.03144700 317.83937 0.000000e+00
## x1 1.0153582 0.03116854 32.57638 4.166280e-159
## x2 0.5096043 0.03093340 16.47424 4.091831e-54Instead of minimizing the error, what if we want to maximize the likelihoods again?
ll.cont.mx <- function(p, X, y){
len <- length(p)
betas <- p[-len]
mu <- X %*% betas
sds <- p[len]
sum(dnorm(y, mean = mu, sd = sds, log = T)) #maximize this
#sum((y - mu)^2) #minimize this
}
start <- ncol(X) + 1 #add one w/c is the variance
fit <- optim(p = rep(1, start), #starting values
fn = ll.cont.mx, #function to use
X = X, y = y, #pass this to our
hessian = T, #to get standard errors
control = list(fnscale = -1, #to maximize
reltol = 1e-10) #change in deviance to converge
)
b <- fit$par[1:3] #the betas
ll <- fit$value #the log likelihood
conv <- fit$convergence #value of zero means convergence
ses <- sqrt(diag(solve(-fit$hessian)))[1:3] #get the SE from the inverse of the Hessian matrix
df <- data.frame(b, ses, t = b/ses)
rownames(df) <- colnames(X)
df #putting it all together ## b ses t
## (Intercept) 9.9951172 0.03139866 318.32944
## x1 1.0153670 0.03112063 32.62681
## x2 0.5095777 0.03088585 16.49874ll #the log likelihood## [1] -1411.179t1 <- lm(y ~ x1 + x2, data = df)
summary(t1)$coef## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 9.9950936 0.03144700 317.83937 0.000000e+00
## x1 1.0153582 0.03116854 32.57638 4.166280e-159
## x2 0.5096043 0.03093340 16.47424 4.091831e-54logLik(t1)## 'log Lik.' -1411.179 (df=4)Similar coefficients, standard errors, and the same log likelihood.
— END