I’ve spoken a bit about how using weights is important when analyzing national/statewide datasets. The weights are used so the sample generalizes to a particular population (note: we are interested in making inferences about the population, not the sample). This is important because at times, in national datasets, certain subpopulations (e.g., Hispanic or Asian students) are oversampled to ensure that the sample size is large enough for subgroup analysis. Without using weights, certain groups may be overrepresented (or underrepresented).
In my own work conducting statewide school climate surveys, principals are provided the option to, in order to lessen the administrative burden, 1) randomly sample 25 students per grade level (i.e., the random sampling [RS] option) or 2) have all students in the school answer the survey (achieve > 80% response rate or the whole grade [WG] option). The probability of selection in each school is different.
Example 1
Consider two schools of 100 students each: In school A, random sampling is used (n = 25) and in school B, the whole grade option is used (n = 100). In school A, the probability of selection is 25/100 = .25. Weights are the inverse of the probability of selection or 1/.25 = 4. In other word, each response is worth four students for school A. For school B, the weights are set to 1 since everyone responded to the survey. So for the entire sample (n = 125), the respondents will have different weights. If a measure of some outcome is taken, the average will be greatly influenced by the school that did the whole grade option [this is a simplified example but you get the point].
Example 2
Take another example. Let’s say we are sampling students in public (N = 1,000) and private schools (N = 10,000). We just randomly sample 100 students in each group (forget the school clustering for now). On average, the students in private school scored 100 on some test and students in public school scored 150. The average of both public and private school students or all students is 125.
| x | Private | Public | Total |
|---|---|---|---|
| Population (N) | 1000 | 10000 | 11000.00 |
| Sample (n) | 100 | 100 | 200.00 |
| Sampling Weight | 10 | 100 | NA |
| Unweighted Mean | 100 | 150 | 125.00 |
| Weighted Mean | 100 | 150 | 145.45 |
However, we know that the sampling rate for each group/strata is different so if we ignore that, we treat all responses equally. In reality, students in private schools had a 100/1000 = 10% chance of being selected. Students in public schools had a 100/10000 = 1% chance. The expansion weight is the inverse of the probability of being selected: for private schools, the weight is \(\frac{1}{.10} = 10\) and for public school students, the sampling weight is \(\frac{1}{.01} = 100\). If we use the weighted mean, the average score is \(145.45\) vs. \(125\) (see below).
The unweighted (regular) mean is \(\bar{x} = \frac{\Sigma x}{n}\).
The weighted mean is \(\bar{x}_{w} = \frac{\Sigma wgt \times x}{\Sigma wgt}\).
To show the difference:
sc <- c(rep(100, 100), rep(150,100))
wt <- c(rep(10,100), rep(100,100))
df <- data.frame(sc, wt)
mean(df$sc) #unweighted## [1] 125weighted.mean(df$sc, df$wt) #weighted mean## [1] 145.4545Example 3
I’ll illustrate using simulated data (this time adding some nesting). I’ll also show how to get some basic weighted descriptive statistics.
Simulate data with weights
library(nlme)
library(lme4)
library(dplyr)
library(jtools)
### can ignore this section, what is important is the dat file at the end
l2s <- 10 #number of clusters
l1 <- 20 #number of units per cluster
G01 <- .3 #effect L2
G11 <- .5 #effect L1
set.seed(12345)
tmp2 <- rnorm(l2s, 0, 1)
tmp3 <- rep(c(0,1), each = 5)
W2 <- rep(tmp2, each = l1) #l2 predictor
FEM <- rbinom(l2s * l1, 1, .5) #gender l1
X1 <- rnorm(l2s * l1, 0, 2) #l1 predictor
e2 <- rep(rnorm(l2s, 0, 1), each = l1) #l2 error
grp <- rep(1:l2s, each = l1)
e1 <- rnorm(l2s * l1, 0, 2) #this controls the ICC
y <- G01 * W2 + G11 * X1 + FEM * .4 + e2 + e1
wts <- rep(c(10, 10, 5, 5, 1), each = 40) #these are the weights
#in this case, just place them after to show differences
#weights could result in: did not use them for the creation of y
dat <- data.frame(y, X1, W2, FEM, grp, wts) #this is our datasetView descriptives
Simple descriptives can be inspected with or without weights:
mean(dat$y)## [1] 0.5997105weighted.mean(dat$y, dat$wts)## [1] 0.8253441#difference of ~0.23 pts
### by gender (FEMale = 1)
dat %>%
group_by(FEM) %>%
summarise(m.u = mean(y),
m.w = weighted.mean(y, wts))## # A tibble: 2 × 3
## FEM m.u m.w
## <int> <dbl> <dbl>
## 1 0 0.252 0.171
## 2 1 0.862 1.31Based on the weights, the differences between weighted and unweighted means increased (.25 vs .86: Unweighted) vs (.17 vs 1.31: Weighted).
Now this can be viewed one variable at time but to simplify this, I can use the tableone package which can give descriptives, by group, using weights and without weights. For unweighted descriptives (stratified by gender):
library(tableone)
vars <- c('y', 'X1', 'W2')
t1 <- CreateTableOne(data = dat, vars = vars, strata = 'FEM')
print(t1, smd = T) #smd = T to show standardized mean differences## Stratified by FEM
## 0 1 p test SMD
## n 86 114
## y (mean (SD)) 0.25 (2.26) 0.86 (2.92) 0.109 0.234
## X1 (mean (SD)) -0.10 (1.96) 0.17 (1.93) 0.327 0.140
## W2 (mean (SD)) -0.07 (0.73) -0.18 (0.81) 0.334 0.139For weighted descriptives (note: requires the survey package to incorporate the weights):
library(survey)
des <- svydesign(ids = ~grp, weights = ~wts, data = dat)
t2 <- svyCreateTableOne(data = des, strata = "FEM", vars = vars)
print(t2, smd = T)## Stratified by FEM
## 0 1 p test SMD
## n 527.00 713.00
## y (mean (SD)) 0.17 (2.28) 1.31 (2.80) 0.051 0.446
## X1 (mean (SD)) -0.18 (1.95) 0.26 (2.07) 0.331 0.219
## W2 (mean (SD)) 0.13 (0.68) -0.04 (0.75) 0.124 0.237We can see greater differences among the variables. This is useful.
Analyze data using multilevel models
[NOTE: THIS SECTION MAY BE INCORRECT– the weights used are NOT sampling weights. Use WeMix instead to get the correct estimates.]
Since we have nested data, we may also analyze the data using a multilevel model. We can begin by viewing the unconditional intraclass correlation coefficient (ICC).
library(lme4)
m0.uw <- lmer(y ~ (1|grp), data = dat) #using lme4
m0.w <- update(m0.uw, weights = wts) #with weights
summary(m0.uw)## Linear mixed model fit by REML ['lmerMod']
## Formula: y ~ (1 | grp)
## Data: dat
##
## REML criterion at convergence: 918
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.56989 -0.60085 0.05237 0.56032 2.70286
##
## Random effects:
## Groups Name Variance Std.Dev.
## grp (Intercept) 2.110 1.453
## Residual 5.201 2.281
## Number of obs: 200, groups: grp, 10
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 0.5997 0.4868 1.232summary(m0.w) ## Linear mixed model fit by REML ['lmerMod']
## Formula: y ~ (1 | grp)
## Data: dat
## Weights: wts
##
## REML criterion at convergence: 977.9
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9805 -0.5211 0.0283 0.4675 3.3282
##
## Random effects:
## Groups Name Variance Std.Dev.
## grp (Intercept) 1.857 1.363
## Residual 34.101 5.840
## Number of obs: 200, groups: grp, 10
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 0.6469 0.4793 1.35Based on the random effects, the unconditional ICC of the unweighted model is .29. For the weighted model, the ICC is .05. That’s a big difference!
Now, if we examine results with and without weights using a full model:
m1.uw <- lmer(y ~ W2 + X1 + FEM + (1|grp), data = dat) #using lme4
m1.w <- update(m1.uw, weights = wts)
#export_summs(m1.uw, m1.w, model.names = c('UnW', 'W'), digits = 3)
library(stargazer)
stargazer(m1.uw, m1.w, type = 'text', digits = 3,
no.space = T, intercept.bottom = F,
star.cutoffs = c(.05, .01, .001),
column.labels = c('UnW', 'W'))##
## ==================================================
## Dependent variable:
## ------------------------------
## y
## UnW W
## (1) (2)
## --------------------------------------------------
## Constant 0.320 0.180
## (0.522) (0.512)
## W2 0.365 0.318
## (0.636) (0.624)
## X1 0.636*** 0.680***
## (0.069) (0.065)
## FEM 0.520 0.768**
## (0.277) (0.268)
## --------------------------------------------------
## Observations 200 200
## Log Likelihood -423.252 -441.384
## Akaike Inf. Crit. 858.505 894.767
## Bayesian Inf. Crit. 878.295 914.557
## ==================================================
## Note: *p<0.05; **p<0.01; ***p<0.001In this case, relationship of gender (FEM = 1) is higher in the weighted version.
Very quick and important sidenote
With R, the two main packages to use when estimating multilevel models are lme4 and nlme. Both are great though researchers should be careful when using weights, especially with nlme. [NOTE: THESE ARE NOT SAMPLING WEIGHTS]
library(nlme)
nlme.u <- lme(y ~ W2 + X1 + FEM, random = ~1|grp, data = dat) #using nlme
nlme.w1 <- update(nlme.u, weights = ~wts) #incorrect
nlme.w2 <- update(nlme.u, weights = ~1/wts) #correct
stargazer(nlme.u, nlme.w1, nlme.w2, m1.w, type = 'text',
digits = 3, no.space = T, intercept.bottom = F,
star.cutoffs = c(.05, .01, .001),
column.labels = c('nlme.UnW', 'nlme.W [wrong]' ,'nlme.W [correct]', 'lmer.W'),
dep.var.caption = '', dep.var.labels.include = F,
model.names = F)##
## =====================================================================
## nlme.UnW nlme.W [wrong] nlme.W [correct] lmer.W
## (1) (2) (3) (4)
## ---------------------------------------------------------------------
## Constant 0.320 0.645 0.180 0.180
## (0.522) (0.539) (0.512) (0.512)
## W2 0.365 0.398 0.318 0.318
## (0.636) (0.648) (0.624) (0.624)
## X1 0.636*** 0.485*** 0.680*** 0.680***
## (0.069) (0.079) (0.065) (0.065)
## FEM 0.520 -0.071 0.768** 0.768**
## (0.277) (0.295) (0.268) (0.268)
## ---------------------------------------------------------------------
## Observations 200 200 200 200
## Log Likelihood -423.252 -476.987 -441.384 -441.384
## Akaike Inf. Crit. 858.505 965.973 894.767 894.767
## Bayesian Inf. Crit. 878.174 985.642 914.436 914.557
## =====================================================================
## Note: *p<0.05; **p<0.01; ***p<0.001Note the way weights are specified. For sampling weights, it has to be written as ~1/weight when using nlme. The results are the same as when lmer was used (see Models 3 & 4; just repeated). It is incorrect to use just ~weight (see Model 2 results w/c are off).
NOTE: this is for functions that use the nlme package. That goes for the glmmPQL function in the MASS package. [WARNING: these are not sampling weights].
If you compare this with what SAS generates using PROC MIXED, you will get the same results.
proc mixed data = a; #imported into dataset a
class grp;
weight wts;
model y = W2 X1 FEM /solution ddfm = kr;
random intercept / subject = grp;
run;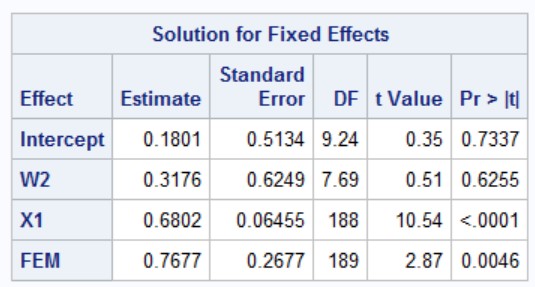
Weighting at two levels (WeMix)
WeMix is a new package for running mixed models with weights at more than one level (the only package that I know of). There is still some debate on how to use weights at the different levels (see Mang et al., 2021).
Testing whether sampling weights are needed
DuMouchel and Duncan (1983 in JASA; Using Sample Survey Weights in Multiple Regression Analyses of Stratified Samples) compared results between weighted and unweighted analyses. Long implemented this test in the jtools package. This works with an lm object. A statistically significant F test suggests that weights are needed. (note: I have never actually used this particular test in my own work but is interesting to know such a test exists). For a review of tests to see if weights are useful, see Bollen et al.’s (2016) article. Basically the two general types of tests are Hausman (1978) tests (i.e., difference in coefficients [DC] tests) and weighted association (WA) tests.
library(jtools)
tt1 <- lm(y ~ W2 + X1 + FEM, data = dat)
weights_tests(model = tt1, weights = wts, data = dat, sims = 100)## DuMouchel-Duncan test of model change with weights
##
## F(4,192) = 6.867
## p = 0
##
## Lower p values indicate greater influence of the weights.
##
## Standard errors: OLS
## ------------------------------------------------
## Est. S.E. t val. p
## ----------------- ------- ------ -------- ------
## (Intercept) 1.18 0.51 2.33 0.02
## W2 2.19 0.56 3.94 0.00
## wts -0.07 0.07 -1.02 0.31
## X1 0.44 0.18 2.48 0.01
## FEM -1.06 0.64 -1.65 0.10
## W2:wts -0.35 0.09 -3.85 0.00
## wts:X1 0.03 0.02 1.19 0.24
## wts:FEM 0.22 0.09 2.38 0.02
## ------------------------------------------------
##
## ---
## Pfeffermann-Sverchkov test of sample weight ignorability
##
## Residual correlation = 0.12, p = 0.05
## Squared residual correlation = -0.10, p = 0.19
## Cubed residual correlation = 0.10, p = 0.12
##
## A significant correlation may indicate biased estimates
## in the unweighted model.References:
Mang, J., Kuchenhoff, H., Meinck, S., & Prenzel, M. (2021). Sampling weights in multilevel modelling: An investigation using PISA sampling structures. Large-Scale Assessments in Education, 9(1), 6. https://doi.org/10.1186/s40536-021-00099-0
#END