Random stuff II: Plotting residuals
I was poking around my old teaching files and I found an old file and I wasn’t sure what it was:
dat <- read.table("https://raw.githubusercontent.com/flh3/pubdata/main/Stefanski_2007/mizzo_1_data_yx1x5.txt")
head(dat)## V1 V2 V3 V4 V5 V6
## 1 -0.22391 0.0054599 0.380310 0.0135140 0.209240 0.1467100
## 2 0.84413 0.1073700 -0.026533 0.0458640 0.012987 -0.0271900
## 3 1.06240 0.0911160 0.181260 0.0501710 -0.188670 -0.0120820
## 4 -1.04170 0.4404900 0.245960 0.0054154 -0.212920 0.1015200
## 5 0.15655 -0.1705100 0.147620 0.0836320 -0.095286 -0.0078451
## 6 -0.13526 0.0616050 -0.804130 -0.0259500 0.291730 -0.0783840dim(dat)## [1] 3785 6Turns out it was an old data file I had used in class discussing regression diagnostics. We often talk about the assumption of the homoskedasticity of residuals and we graphically depict that by plotting the fitted values on the X axis and the residuals on the y axis. If all is well, we are told that we should have any discernible pattern.
So this is a dataset of 3,785 observations and 6 variables. We can predict the first variable (V1) using all the other variables in the dataset (V2 to V6).
m1 <- lm(V1 ~ ., data = dat)If we plot the residuals, we get:
plot(fitted(m1), resid(m1))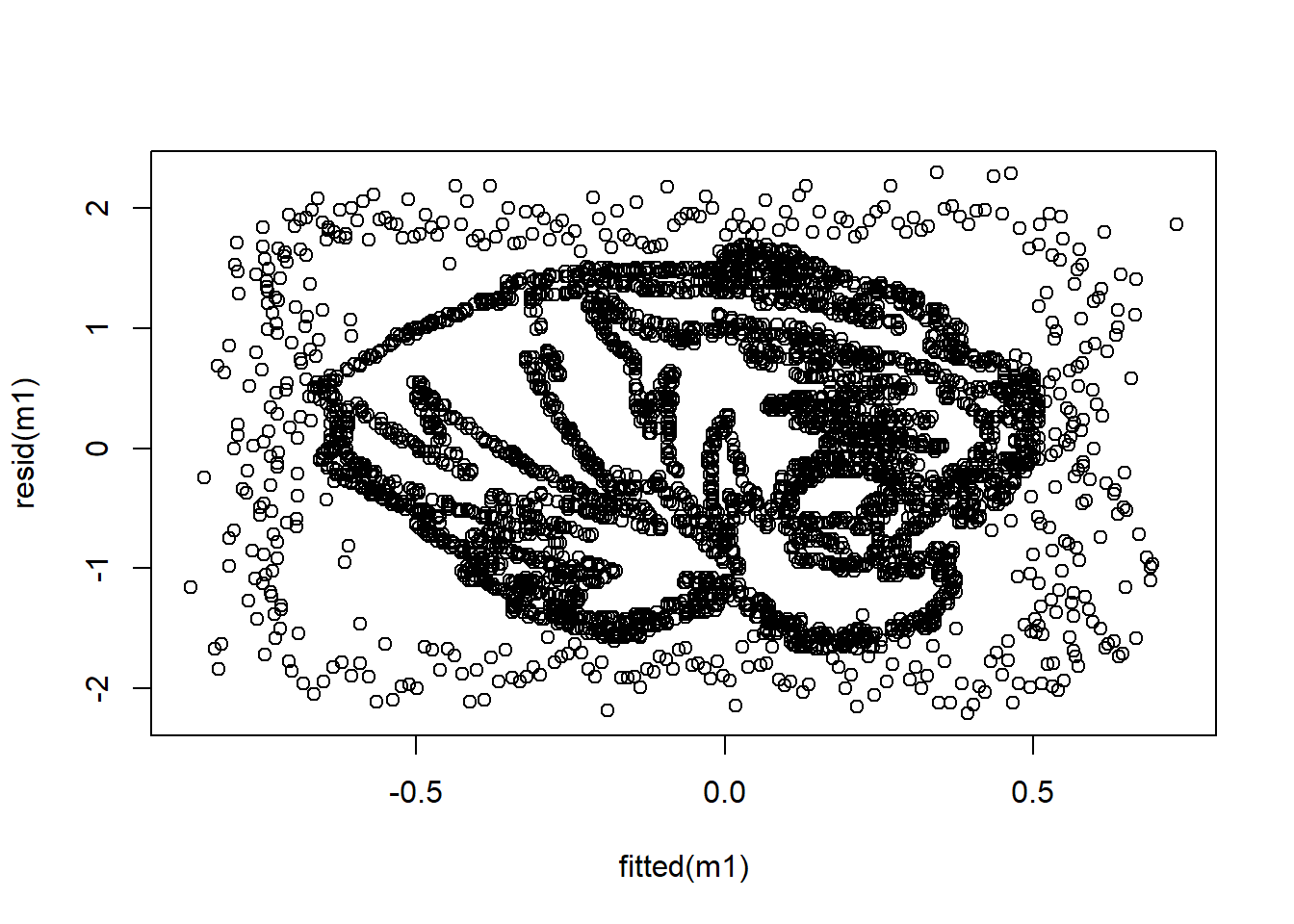
Just thought that was neat. This is based on the work of:
Stefanski, L. A. (2007). Residual (sur)realism. The American Statistician, 61(2), 163-177. https://doi.org/10.1198/000313007X190079
I can’t find the original website where this came from but definitely check out the paper!
Here’s the original image:

Figure 1: MU TIGERS
– END